Normalized Difference Indices#
Under Construction.
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
import netCDF4 as nc4
Overview#
Under Construction.
Physical Basis#
Under Construction.
# read in the data
sand_reflectance = np.genfromtxt('../data/sand_reflectance_spectra.csv', delimiter=',')
leaf_reflectance = np.genfromtxt('../data/leaf_reflectance_spectrum.txt', delimiter=',')
# make a figure
plt.figure(figsize=(8,4))
plt.gca().add_patch(Rectangle([0.64,0],0.03,60,facecolor='silver',alpha=0.5))
plt.gca().add_patch(Rectangle([0.85,0],0.03,60,facecolor='silver',alpha=0.5))
plt.plot(sand_reflectance[:,0]*1e-3, sand_reflectance[:,1]*100, color='brown')
plt.plot(leaf_reflectance[:,0], leaf_reflectance[:,1], color='forestgreen',
label='Big Bluestem Grass')
plt.gca().set_xlim([0.52,0.9])
plt.gca().set_ylim([0,60])
plt.ylabel('Reflectance')
plt.xlabel('Wavelength ($\mu$m)')
plt.legend()
plt.show()
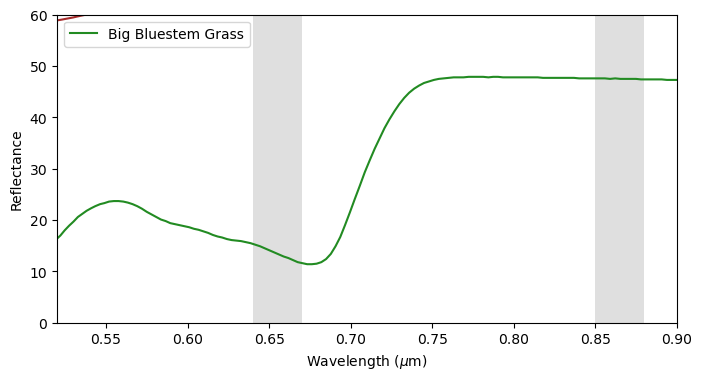
The soil reflectance data is obtained from HUEMMRICH [1994]. The leaf reflectanc data is obtained from MIDDLETON [1994].
An Example of Common Indices#
To demonstrate some of the common indices, we can look at several bands from a Landsat 9 of Mount Taranaki, New Zealand, taken on 27 June 2024. Mount Taranaki is a unique location in that it is a large, snow-capped volcano surrounded by a vegetation region on the southwest coast of New Zealand’s north island. Here, the scene has been cropped to the region around Mount Taranaki and subsetted to bands 2-6. We can read this data in as follows:
# read in the data
ds = nc4.Dataset('../data/taranaki_landsat.nc')
band_2 = ds.variables['band_2'][:, :]
band_3 = ds.variables['band_3'][:, :]
band_4 = ds.variables['band_4'][:, :]
band_5 = ds.variables['band_5'][:, :]
band_6 = ds.variables['band_6'][:, :]
ds.close()
Before computing some normalized difference indices for this scene, let’s take a look at a “Natural Color” image of this scene to get a feel for some components of the image:
# make an RGB array
RGB = np.stack([band_4, band_3, band_2], axis=-1)
# clip high and low values to show colors in range
minval = np.percentile(RGB, 2)
maxval = np.percentile(RGB, 98)
RGB = np.clip(RGB, minval, maxval)
# rescale so that numbers are 8 bit
RGB = ((RGB - minval) / (maxval - minval)) * 255
RGB = RGB.astype(int)
# make a figure
plt.figure()
plt.imshow(RGB)
plt.axis('off')
plt.title('"Natural Color" Image of Mount Taranaki')
plt.show()
/Users/mike/opt/anaconda3/envs/ms274/lib/python3.10/site-packages/numpy/lib/function_base.py:4823: UserWarning: Warning: 'partition' will ignore the 'mask' of the MaskedArray.
arr.partition(

As we can see, this scene has Mount Taranaki in the bottom right of the image. It has a snow-capped peak in the middle and is surrounded by dark vegetation on the volano slopes. Around these locations, there is a different type of vegetation, shown by the light green color. In the left and upper part of the image, we can see the coastal ocean. Finally, we can see some clouds in the scene in the upper left corner of the image and on the lower-right side of the volcano. In effect, this scene allows us to test the indices for vegetation (NDVI), water (NDWI), and snow (NDSI).
NDVI#
# make the NDVI array
NDVI = (band_5-band_4)/(band_5+band_4)
# make a plot
plt.figure()
C = plt.imshow(NDVI, vmin=-0.5, vmax=0.5, cmap='BrBG')
plt.colorbar(C, label='NDVI')
plt.axis('off')
plt.show()
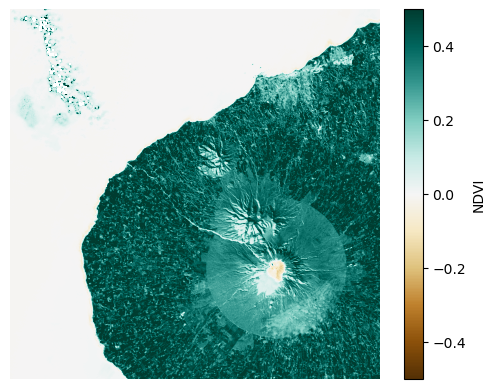
Here, we can see that the vegetated region around the volcano has very high values of NDVI relative to the water in the ocean and the snow on the top of the mountain.
NDWI#
# make the NDWI array
NDWI = (band_3-band_5)/(band_3+band_5)
# make a plot
plt.figure()
C = plt.imshow(NDWI, vmin=-0.25, vmax=0.25, cmap='RdBu')
plt.colorbar(C, label='NDWI')
plt.axis('off')
plt.show()
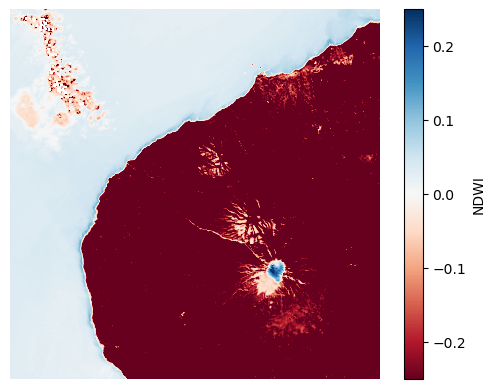
In this scene, we see positive values of NDWI in the ocean and in the snow on top of the mountain, and negative values in the clouds and on the vegetation.
NDSI#
# make the NDSI array
NDSI = (band_3-band_6)/(band_3+band_6)
# make a plot
plt.figure()
C = plt.imshow(NDSI, vmin=-0.5, vmax=0.5, cmap='RdBu')
plt.colorbar(C, label='NDSI')
plt.axis('off')
plt.show()
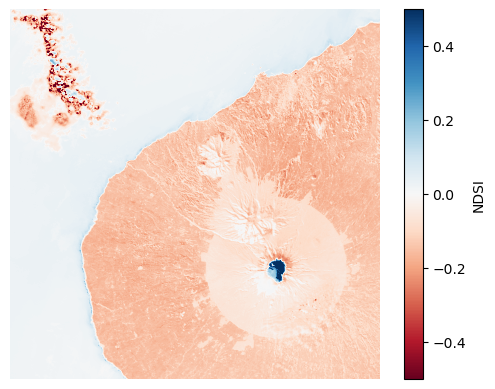
Finally, for NDSI, we see the highest values on the top of the mountain relative to anywhere else on the scene.
